We are looking for DBBS/MSTP/CS/MATH/EE/BME students to rotate in our lab!
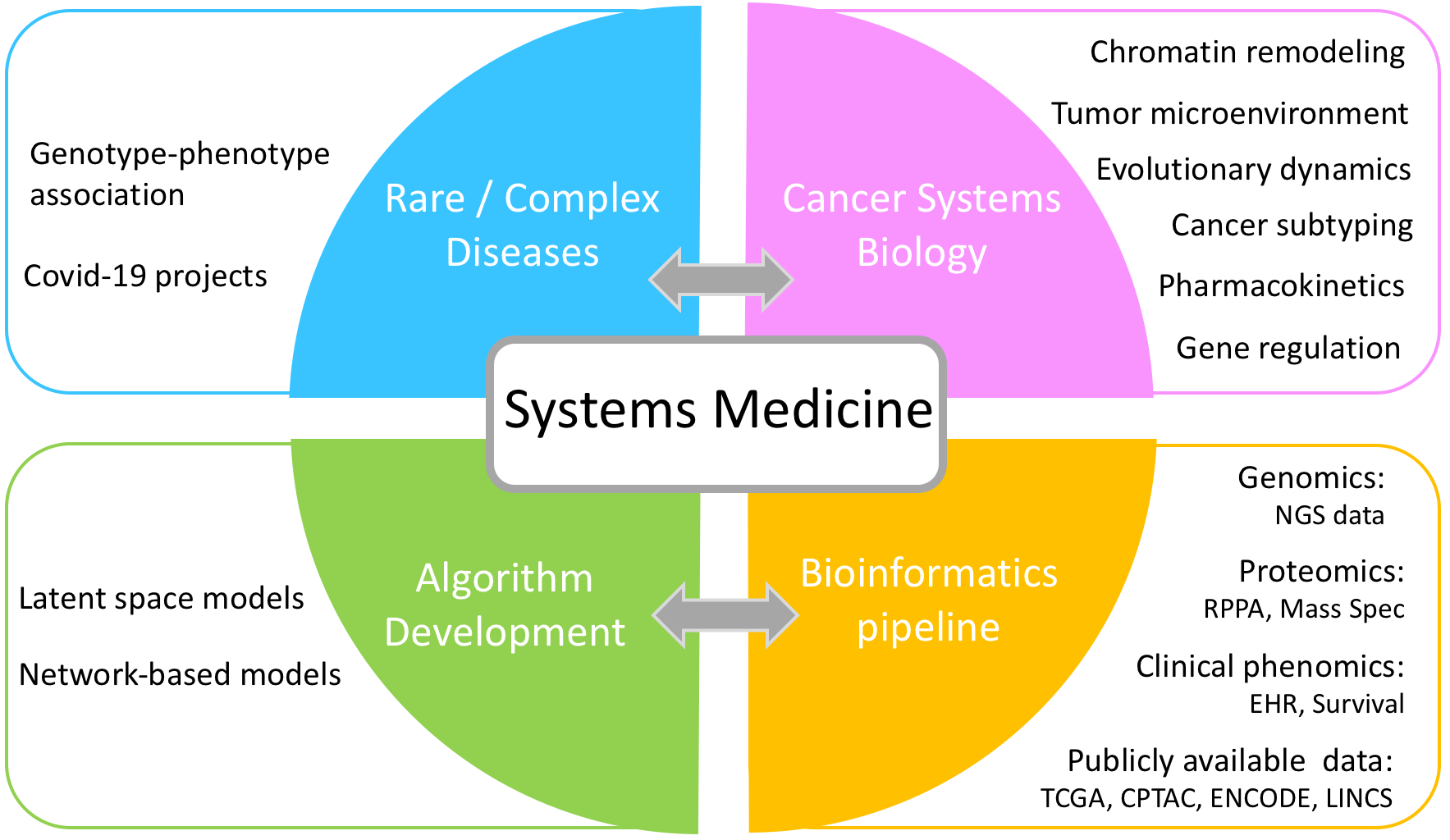
Our research goal is to interpret and distill the complexity of cancer and other rare diseases through integration of large scale multi-omics data using dynamic modeling, graph theory, and machine learning methods. We aim to apply these methods to study challenging cancer biology problems, particularly how chromatin alterations influence cellular phenotypes in response to genetics, environments, and pharmacological perturbations. By integrating large datasets, we hope to extract relevant information necessary to make precise biological and clinical predictions and computationally direct experiments. The primary focus of our lab is to produce high-resolution computational models to study the effects of genetic and epigenetic perturbations on chromatin alterations that affect cellular states, elucidating the molecular mechanisms of cancer and other diseases.
Mission statement: Spread awareness of and inspire involvement in STEM through mentorship and outreach efforts both on and off-campus. Our lab is committed to helping students in their careers and become the best version of themselves by achieving their goals, introducing them to critical thinking, challenging their limiting assumptions, teaching them life lessons, and much more. We strive to cross-pollinate students from various STEM disciplines toward utilizing computational biology and systems approaches to address the underlying disease mechanisms. We are also committed to empowering and inspiring next-generation women in STEM field.
Research Projects
Cancer systems biology
— Chromatin remodeling in cancer (histone modifications)
— Cancer subtyping
— Tumor microenvironment
— Targeting cancer stemness pathway in breast cancer
— Single-cell approaches to address tumor heterogeneity
— Pharmacodynamics & pharmacokinetics of anti-cancer drugs
— Biomarker discovery in cancer
— Cancer immunotherapy
- Modeling gene regulatory networks
- Genotype-phenotype correlation
- Natural language processing
